|

In 1963, a decade after the Watson and Crick model of DNA was proposed, the startling discovery of a new form of DNA was made by John Cairns of Australian National University- that of DNA rings. In DNA rings, the two strands of the double helix wind around each other and they form a pair of inter-twined closed curves in space.
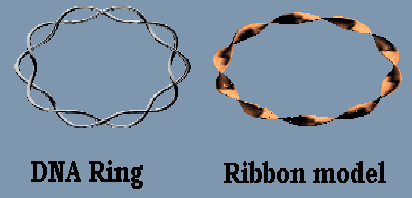
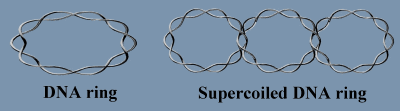
In the micrographs of the DNA rings, some of the rings were supercoiled, i.e., the axis of the double helix is itself coiled up in the form of a helix.
The biochemical properties alone of DNA could not adequately explain this phenomenon. Ignoring these properties and looking at the DNA molecules as mathematical objects (curves in space, ribbons) provides a solution to the problem. Techniques of Topology and Differential Geometry are used.
To study the closed circular configuration of the DNA molecule mathematically, it is most convenient to construct a model in which the structure is represented as a narrow twisted ribbon. We construct this model from the twisted ladder model of DNA.
The edges of the ribbon represent the two strands and the axis of the double helix is mapped onto the line that bisects the ribbon. When the two ends of the ribbon are joined together, each edge describes a closed curve in the three dimensional space. The ribbon forms a ring which twists as the double helix twists and its axis supercoils as the axis of the double helix supercoils in space.
The model is analyzed in three different ways. This gives us three different quantities- Twist, Link and Writhe.
Twist
Now, we look at the whole ribbon. Twist tells us the number of complete turns that exist in the ring formed by the ribbon.
Copyright © 2022 ICICI Centre for Mathematical Sciences
All rights reserved. Send us your suggestions at .
|